Jun 11, 2025
TUNEL protocol that preserves protein antigenicity for co- or iterative immunofluorescence (e.g. MILAN, CycIF)
- Marc S Sherman1,2,
- Thomas Skates1,
- Lindsey Gaston3,2,
- Sonya Katzen1,
- joseph.majzoub 3,2,
- Wolfram Goessling1,2
- 1Massachusetts General Hospital;
- 2Harvard Medical School;
- 3Boston Children's Hospital

Protocol Citation: Marc S Sherman, Thomas Skates, Lindsey Gaston, Sonya Katzen, joseph.majzoub , Wolfram Goessling 2025. TUNEL protocol that preserves protein antigenicity for co- or iterative immunofluorescence (e.g. MILAN, CycIF). protocols.io https://dx.doi.org/10.17504/protocols.io.261ge5327g47/v1
License: This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Protocol status: In development
We are still developing and optimizing this protocol
Created: May 16, 2024
Last Modified: June 11, 2025
Protocol Integer ID: 99954
Keywords: TUNEL, MILAN, CycIF, cell death, cyclic immunofluorescence, Multiple Iterative Labeling by Antibody Neodeposition, apoptosis, necrosis, Terminal deoxynucleotidyl transferase dUTP nick end labeling, downstream iterative immunofluorescence, iterative immunofluorescence, protein antigenicity, antigen retrieval, incompatibilities between tunel, tunel, immunofluorescence
Funders Acknowledgements:
Marc Sherman
Grant ID: K08DK139370
Abstract
TUNEL is not known to be compatible with modern multiplexed, iterative immunofluorescence. Here we demonstrate a variation of TUNEL which resolves identified incompatibilities between TUNEL and both MILAN and CycIF. The protocol involves replacing proteinase K digestion with a pressure cooker induced antigen retrieval. Expected results are identical to having conducted TUNEL using proteinase K - but completely prevent (and in fact, enhance) protein antigenicity in concurrent and downstream iterative immunofluorescence.
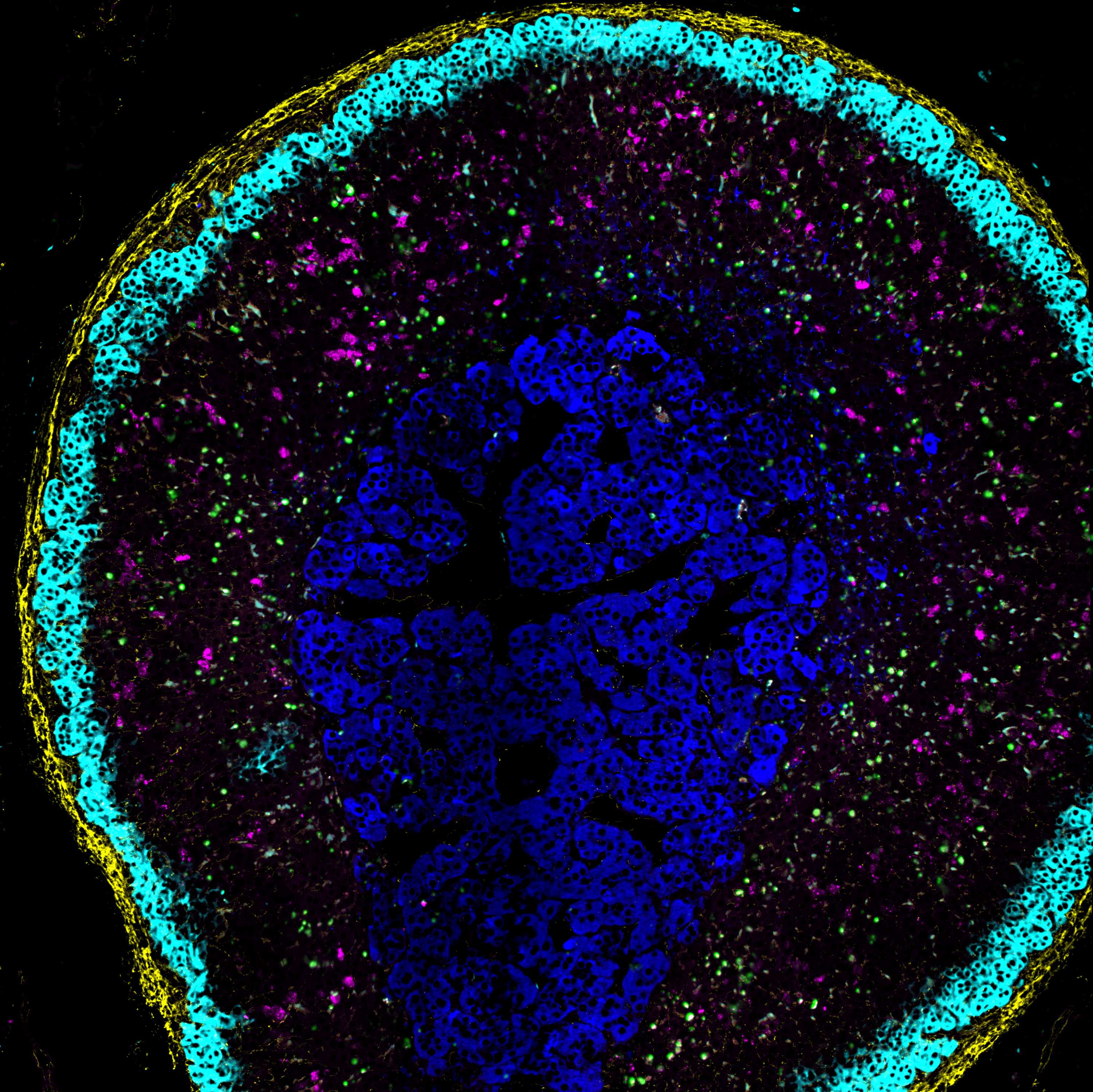
Image Attribution
Adrenal gland during dexamethasone suppression induces marked apoptosis.
Guidelines
This protocol details how to perform TUNEL while maintaining (bidirectional) compatibility with a highly multiplexed and/or iterative immunofluorescence staining series as might be done with MILAN or CycIF. The optimal order is to perform TUNEL first, however, we show TUNEL may work at least 4 rounds into a MILAN staining series as well. The protocol below is "TUNEL-first", in part to highlight the antigen retrieval method. Please see variations below for how to incorporate this protocol at later stages.
Conceptually, the primary two incompatibilities of conventional TUNEL with highly multiplexed IF--or even conventional IF--are the following.
1. Proteinase K digestion must not be performed, or risk loss of antigenicity for antibody-based immunofluorescence which ranges from modest to complete.
2. Use of pressure cooker is required; pressure cooking 'rescues' TUNEL activity while also enhancing protein antigenicity in IF.
Steps that probably don't matter:
- the permeabilization buffer (we use MILAN wash - but likely any classic permeabilization buffer should work)
- the precise deparaffinization steps; ours is optimized for liver tissue, worked for adrenal, breast cancer, and mammary tissue, but variations are also likely to work as long as they do not involve drying the tissue
- the precise photobleaching apparatus is likely not important, as long as wavelengths that might induce DNA damage (and therefore false TUNEL signal) are avoided (e.g., significant UV light exposure)
Variations:
1. TUNEL with protein-preserving co-immunofluorescence (ie, no iterative staining). Although the goal of this project was to harmonize TUNEL with an iterative immunofluorescence method like MILAN, the protocol also serves as a protein/antigen-sparing TUNEL method for co-immunofluorescence. This is precisely what is offered by commercial fluorescent-based TUNEL kits, but cheaper (only requires the NEB terminal transferase enzyme kit), easier (removes proK, and an extra fixation step), and retains the benefit of preserved protein antigenicity which commercial ProK-containing kits do not. To do conventional TUNEL with co-immunofluorescence, enter your preferred (FFPE-based) immunofluorescence protocol immediately following the TUNEL reaction (after step 6.7).
2. TUNEL in the middle of a MILAN staining series. Before starting the MILAN series, samples must undergo pressure cooker antigen retrieval. Then proceed with MILAN per usual. At the round where TUNEL is desired, finish stripping, wash in PBS x3, and then enter at step 6.
3. Other iterative immunofluorescence methods (e.g., CODEX/phenocycler, 4i, imaging mass spec). We suspect performing TUNEL first should be compatible with these methods, as the biochemical principles are the same as CycIF and MILAN. One would need to verify that the BrdU incorporation can be read out; for 4i, this is straightforward, but for phenocycler and imaging mass spec, DNA barcoded or heavy metal (respectively) conjugated anti-BrdU antibodies would need to be validated.
4. Proteinase K-based TUNEL with erasure. We show in the accompanying manuscript that proteinase K does not influence erasure; thus, traditionalists may choose to perform TUNEL using proteinase K, and that is still compatible with iterative immunofluorescence excepting the loss of antigenicity. Replace the pressure cooker step with tissue-determined proteinase K digestion followed by 5 minutes of formalin fixation and washes.
Materials
- Xylene
- Ethanol
- PBS
- Platinum LED P300 (300w) grow lamp or equivalent
- Milan Wash Buffer
- DAPI Soft Mount
Protocol materials
CELLSTAR OneWell Plategreiner bio-oneCatalog #670102
ImmEdge hydrophobic barrier pap penVector LaboratoriesCatalog #H-4000
Anti-BrdU antibodyAbcamCatalog #ab152095
BrdU Monoclonal Antibody (MoBU-1)Thermo Fisher ScientificCatalog #B35128
Anti-BrdU antibody [BU1/75 (ICR1)]AbcamCatalog #ab6326
Terminal Transferase - 500 unitsNew England BiolabsCatalog #M0315S
BrdUTP (5-Bromo-2´-Deoxyuridine 5´-Triphosphate), 10 mM in TE bufferThermo FisherCatalog #B21550
DNAse IRocheCatalog #04716728001
Troubleshooting
Before start
Plan out the entire MILAN or CycIF series - when to do background staining, what primaries and secondaries are in each round, etc. If you have never done MILAN or CycIF, we would recommend becoming proficient with one of these methods first. Steps like soft-mounting the coverslips and how to decoverslip, imaging the exact same ROIs each round, and the computational registration require training and experience. In contrast, once facile with an iterative immunofluorescence method, adding in TUNEL will be straightforward.
Deparaffinization - rehydrate sections (1st time only)
45m
Immerse the slides in xylene 2 times for 00:10:00 each
10m
Immerse the slides in 100% Ethanol 2 times for 00:10:00 each
10m
Immerse the slides in 95% Ethanol for 00:05:00 each
5m
Immerse the slides in 70% Ethanol 00:05:00 each
5m
Immerse the slides in 50% Ethanol 00:05:00 each
5m
Rehydrate the slides in 1X PBS for 00:10:00
10m
Photobleaching (optional, for high autofluorescence tissue | 1st time only)
Submerge slides in container with PBS. Lay flat with tissue facing up inside a one-well plate. CELLSTAR OneWell Plategreiner bio-oneCatalog #670102
Turn on light source for time determined experimentally for tissue type and light source.
Note
NOTE 1: For mouse and human liver, we photobleach for 8 hours. We recommend this helpful reference by Duong and Han.
Citation
LINK
NOTE 2: If light source produces heat, make sure to keep the container with the PBS cold by surrounding with ice or placing on a cooling element.
NOTE 3: We also used the CycIF photobleaching setup, 1h of white light, omitting H2O2 in the first round of photobleaching, and can confirm compatibility. As H2O2 may induce DNA damage, we have not evaluated the standard CycIF photobleaching buffer prior to TUNEL.
Pressure cooker-induced antigen retrieval (1st time only)
55m
Immerse slides in TE (pH=9) buffer in a staining bucket, and place inside the pressure cooker basin; add 500mL H2O. Set for 20 minutes at pressure. Total time on this machine is 00:45:00
Equipment
Digital Decloaking Chamber
NAME
Pressure cooker
TYPE
Biocare Medical
BRAND
DC2002
SKU
LINK
45m
Once depressurized, remove using hot gloves. Let cool to 50C (~12-15min) on the bench.
Wash slides in water for 00:05:00
5m
Wash slides in PBS for 00:05:00
Note
Slides may be stored in PBS overnight at 4 °C , or possibly longer. Beyond 1-2 days, recommend adding sodium azide (0.1%, final concentration) to PBS to prevent bacterial growth.
5m
Permeabilization
10m
Wash and permeabilize the slides in MILAN wash buffer for 00:10:00
10m
Wash the slides 2 times in 1X PBS for 00:05:00
5m
Draw a hydrophobic barrier around the sections on each slide and return the slides to 1X PBS
ImmEdge hydrophobic barrier pap penVector LaboratoriesCatalog #H-4000
Background imaging (optional)
3m
Coverslip with DAPI for background imaging. De-coverslip as per MILAN or CycIF directions. Wash 3x in PBS for 00:03:00 each after decoverslipping.
3m
TdT (TUNEL) reaction (A-tailing with BrdUTP)
1h 43m
Preheat incubator to 37 °C
Make TdT reaction mastermix. Add enzyme last; keep mastermix on ice.
| A | B | C | |
| # slides | 1 | 5 | |
| TdT buffer | 5uL | 25uL | |
| CoCl2 | 5uL | 25uL | |
| BrdUTP (10mM) | 0.5uL | 2.5uL | |
| TdT (terminal transferase) | 0.5uL | 2.5uL | |
| ddH2O | 39uL | 195uL | |
| TOTAL | 50uL | 250uL |
Terminal Transferase - 500 unitsNew England BiolabsCatalog #M0315S
BrdUTP (5-Bromo-2´-Deoxyuridine 5´-Triphosphate), 10 mM in TE bufferThermo FisherCatalog #B21550
Note
Critical step. Perform the following controls with a TUNEL reaction:
1. No-BrdUTP (negative control). Omit BrdUTP from the cocktail above, but otherwise follow all steps of the protocol. Expected result: no TUNEL signal in the expected channel.
2. No primary (negative control). Include all reagents, but leave out the primary antibody in CycIF or MILAN. Expected result: no TUNEL signal in the expected channel. *We often skip this control in favor of background imaging, which is functionally a no-primary step, once we have validated the reagents & protocol are working and that there is not substantial secondary antibody non-specific staining.
3. DNase (positive control). Prior to TUNEL reaction (step 4.1), add 1U DNAse IRocheCatalog #04716728001 plus appropriate 10X Dnase buffer (comes with kit), enough volume to cover the section, incubate at 37C for 10 minutes, then wash 3X in PBS and proceed with TUNEL reaction.
Remove slides from 1X PBS and tap the side of each slide on a paper towel
Add 50 µL of the TdT reaction master mix to each section (move quickly and only process one slide at a time to avoid to drying out samples).
Gently place a small square of paraffin on top of the section to evenly coat the section in the reaction mix. Make sure no bubbles form on top of the tissue.
Incubate for 01:30:00 at 37 °C in a dark humidified slide box (place wet paper towel in basin of slide box for humidified slide box)
1h 30m
Rinse slides 3x in 1X PBS baths for 00:03:00
3m
Proceed with MILAN or CycIF (including anti-BrdU in this round of immunostaining).
MILAN1 step
Perform MILAN - Multiple Iterative Labeling by Antibody Neodeposition
Proceed immediately to "Step 4: IF STAINING: Primary and Secondary Ab dilution and incubation" in the Multiple Iterative Labeling by Antibody Neodeposition (MILAN) v5 protocol found here.
To detect the TUNEL reaction from step 4, use an anti-BrdU primary antibody. Here are antibodies and dilutions we have successfully used:
Anti-BrdU antibodyAbcamCatalog #ab152095 dilute 1:1000; choose appropriate anti-rabbit secondary in chosen channel
BrdU Monoclonal Antibody (MoBU-1)Thermo Fisher ScientificCatalog #B35128 dilute 1:100; choose appropriate anti-IgG1 isotype specific secondary in chosen channel
Anti-BrdU antibody [BU1/75 (ICR1)]AbcamCatalog #ab6326 dilute 1:200; choose appropriate anti-rat secondary in chosen channel
Note
As the anti-BrdU stains quite intensely, we perform the first stripping in SDS/2-ME in MILAN for 2 hours shaking instead of the recommended 30 minutes (see Step 5: Stripping in the MILAN protocol); we have not verified whether a shorter erasure length may be sufficient.
Protocol references
Cattoretti G, Bosisio FM, Marcelis L, Bolognesi MM. Multiple iterative labeling by antibody neodeposition (Milan). Published online September 25, 2019. doi:10.21203/rs.2.1646/v5
Bolognesi, M. M. et al. Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. J Histochem Cytochem 65, 431–444 (2017).
Jia Ren Lin, Benjamin Izar, Zoltan Maliga, Yu-An Chen, Giorgio Gaglia, Ziming Du, Clarence Yapp, Shaolin Mei, Sandro Santagata, Peter Sorger 2020. Tissue Cyclic Immunofluorescence (t-CyCIF). protocols.io
Muhlich JL, Chen Y, Yapp C, Russell D, Santagata S, Sorger PK, Stitching and registering highly multiplexed whole-slide images of tissues and tumors using ASHLAR. Bioinformatics 38(19). doi: 10.1093/bioinformatics/btac544
Lin, J.-R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018).
Citations
Step 2.1
Duong H, Han M. A multispectral LED array for the reduction of background autofluorescence in brain tissue.
https://doi.org/10.1016/j.jneumeth.2013.08.018