May 25, 2020
- Bogdan Budnik1,
- Ezra Levy1,
- Guillaume Harmange1,
- Nikolai Slavov1
- 1Northeastern University
- Metabolomics Protocols & WorkflowsTech. support email: bbmisraccb@gmail.com

External link: http://scope.slavovlab.net/
Protocol Citation: Bogdan Budnik, Ezra Levy, Guillaume Harmange, Nikolai Slavov 2020. SCoPE-MS. protocols.io https://dx.doi.org/10.17504/protocols.io.bgt4jwqw
License: This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Protocol status: Working
We use this protocol and it's working
Created: May 25, 2020
Last Modified: May 25, 2020
Protocol Integer ID: 37468
Keywords: single cell proteomics by mass spectrometry, single cell proteomic, proteome configurations of single cell, protein levels in single cell, cell proteome, ms cellular heterogeneity, measuring proteome configuration, single cell, distinct human cancer cell type, proteome, differentiating mouse embryonic stem, distinct regulatory patterns at the mrna, transcriptome, mouse embryonic stem, mass spectrometry, protein covariation, quantifying protein level, many gene, cell population, protein abundance relationship, coordinated mrna, developmental gene, gene, functional phenotype, cell type, cell
Abstract
Cellular heterogeneity is important to biological processes, including cancer and development. However, proteome heterogeneity is largely unexplored because of the limitations of existing methods for quantifying protein levels in single cells. To alleviate these limitations, we developed Single Cell ProtEomics by Mass Spectrometry (SCoPE-MS), and validated its ability to identify distinct human cancer cell types based on their proteomes. We used SCoPE-MS to quantify over a thousand proteins in differentiating mouse embryonic stem (ES) cells. The single-cell proteomes enabled us to deconstruct cell populations and infer protein abundance relationships. Comparison between single-cell proteomes and transcriptomes indicated coordinated mRNA and protein covariation. Yet many genes exhibited functionally concerted and distinct regulatory patterns at the mRNA and the protein levels, suggesting that post-transcriptional regulatory mechanisms contribute to proteome remodeling during lineage specification, especially for developmental genes. SCoPE-MS is broadly applicable to measuring proteome configurations of single cells and linking them to functional phenotypes, such as cell type and differentiation potentials.
Troubleshooting
Plate preperation
Put 1ul of HPLC grade water into each well of a plate using the Nanodrop or a multi-channel pipette.
Conver up the plate and freeze until sorting.
Cell Prep
For suspension cells simply place 1 to 2mL of cell culture in an Eppendorf tube and start step 8. For Adherent cells start at step 4.
Aspirate media off of plate
Wash with 2mL of PBS at 37C 2ml for a 6mm plate and 4ml for a 10mm plate
Add 1ml of Acutase for a 6mm plate and 2ml for a 10mm plate, and incubate at 37 C for 5 minutes
Pipette Acutase around the plate to wash off all the cells and place in a 2mL Eppendorf tube
Spin down cells at 4C at 500 rcf for 2 minutes
Remove supernatant without disturbing the pellet, and re-suspend in PBS at 4C
Spin down cells at 4C at 500 rcf for 2 minutes
Remove supernatant without disturbing the pellet, and re-suspend in PBS at 4C
Cell sorting
Sort cells using teh FACS sorter.
*protocol for this can be found here: https://www.protocols.io/view/aria-plate-sorting-j4kcquw
*ensure that there is a bulk carrier that can be used for the cells sorted, or else you musk make your own
Flash-pop Lysis
Place place the plate into the -80 for at least 5 minutes
00:05:00
Turn on the HEATLYSE protocol (90C for 10 minutes) on the thermocycler and hit pause
Once the thermocycler has hit 90C and at least 5 minutes in the -80 have passed, put the plate into the thermocycler and hit resume (thermocycler will hold 90C for 10 minutes and then go to 4C)
00:10:00
Digestion and Labeling
Once the thermocycler has gone down to 4C remove the plate and spin it down.
If doing 1 plate, make 120ul of a 200mM TEAB, 10ng/ul trypsin solution. Dispense 1ul of of this solution to each well. (Goal here is to have a 2ul reaction with a final concentration of 100mM TEAB and 10ng/ul Promega trypsin gold).
Run the DIGEST protocol on the thermocycler (37C or 45C for 3 hours depending on trypsin used and then goes down to 4C)
03:00:00
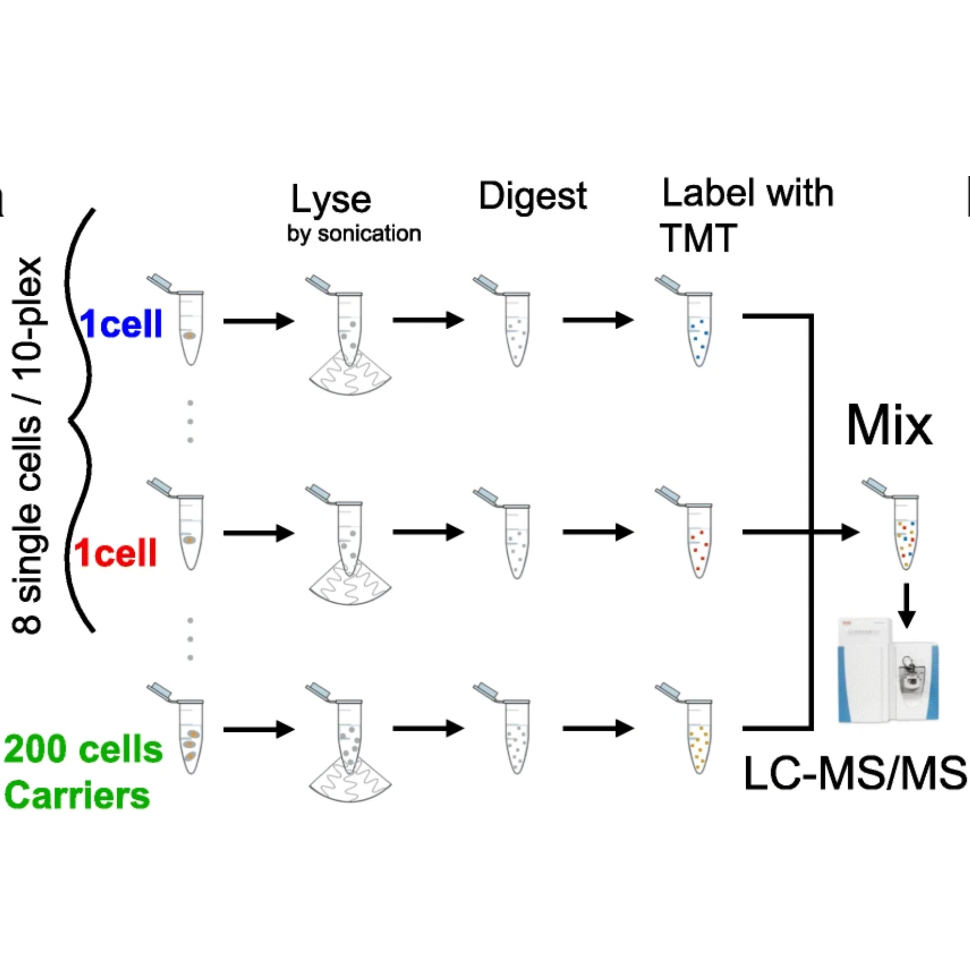
Once the digest is complete, add 0.5 ul of the appropriate 85mM TMT label to each single cell well (Single cells: 127N, 128N, 128C, 129N,129C ,130N, 130C, 131N, 131C, Carrier: 126, Reference: 127C)
Let samples incubate for 1 hour at room temp (22C)
01:00:00
Add 0.5 ul of 0.5% hydroxylamine to all samples, and let incubate at room temp for 30 min.
00:30:00
Submitting to MS/MS
Add 1ul of bulk carrier (contains 100cells worth of material labled 126 and 5 cells of material labeled as 127C) along with 0.5ul of 20% FA to one of the wells in the set.
Aspirate all of the contents of the well where the carrier and FA was added and transfer it to the next well in the set.
Keep aspirating everything in the well dispensing it in the next well aspirating everyting and moving onto the next well until the entire set is in the pippette tip.
Empty all this material in the pippette tip into an MS glass insert, and add water or evaporate liquid so that 12ul are left in the tube.
Place the insert in a labled MS vial and cap the vial with a pierceable cap
Submit to QE using SC method, injecting 10 ul
