Feb 27, 2020
Version 1
sciMAP-ATAC V.1
- Andrew Adey1,
- Casey Thornton1
- 1Oregon Health & Science University

External link: doi: https://doi.org/10.1101/407668
Protocol Citation: Andrew Adey, Casey Thornton 2020. sciMAP-ATAC. protocols.io https://dx.doi.org/10.17504/protocols.io.5r4g58w
License: This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Protocol status: In development
We are still developing and optimizing this protocol
Created: July 24, 2019
Last Modified: February 27, 2020
Protocol Integer ID: 26140
Keywords: Genomics, Epigenomics, Single Cell, Biotechnology, single cell profiling of chromatin state, throughput single cell genomic assay, single cell genomic assay, resolved single cell profiling, cell combinatorial indexing on microbiopsy, cell atac, accessible chromatin, transposase accessible chromatin, chromatin state, cell localization, cell combinatorial indexing, heterogeneity of cell state, cell data, understanding complex tissue, cell data of equivalent quality, interconnected cell, high density multiregional sampling, cell state, complex tissue, cell
Abstract
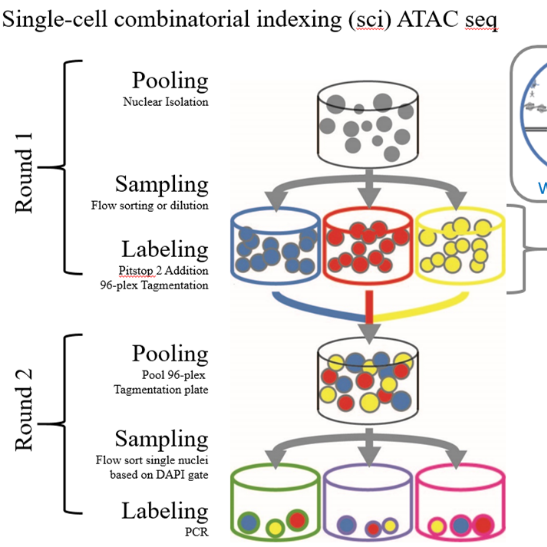
High-throughput single cell genomic assays resolve the heterogeneity of cell states in complex tissues, however, the spatial orientation within the network of interconnected cells is lost. As cell localization is a necessary dimension in understanding complex tissues and disease states, we present a tool for highly scalable spatially-resolved single cell profiling of chromatin state. We use high density multiregional sampling to perform single-cell combinatorial indexing on Microbiopsies Assigned to Positions for the Assay for Transposase Accessible Chromatin (sciMAP-ATAC) to produce single-cell data of equivalent quality to non-spatial single-cell ATAC-seq.
Materials
MATERIALS
Magnesium ChlorideFisher ScientificCatalog #AC223210010
IGEPAL-CA630Merck MilliporeSigma (Sigma-Aldrich)Catalog #I3021 SIGMA-ALDRICH
Triton X-100Merck MilliporeSigma (Sigma-Aldrich)Catalog #T8787-50ML
Tween-20Merck MilliporeSigma (Sigma-Aldrich)Catalog #P-7949
Sodium ChlorideFisher ScientificCatalog #S271-3
Agencourt Ampure XPBeckman CoulterCatalog #A63880
4,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI)Thermo Fisher ScientificCatalog #D1306
Embedding base moldsFisher ScientificCatalog #22-363-553
Jung tissue freezing medium (Leica Microsystems) or OCT compound (TissueTek)
Cell strainer, 35 μm CorningCatalog #352235
Pierce Preotease Inhibitor Tablets, EDTA-FreeThermo Fisher ScientificCatalog #A32955
Tris-HClLife TechnologiesCatalog #AM9855
Superfrost Plus Microscope SliesThermo Fisher ScientificCatalog #4951PLUS4
1X PBS, cell culture gradeThermo Fisher Scientific
Potassium ChlorideMerck MilliporeSigma (Sigma-Aldrich)Catalog #P9541
EDTAInvitrogen - Thermo FisherCatalog #AM9261
Qiagen ProteaseFisher ScientificCatalog #NC9221823
Pitstop 2Merck MilliporeSigma (Sigma-Aldrich)Catalog #SML1169-5MG
Nextera DNA Flex Library PrepIllumina, Inc.Catalog #20018705
QIAquick PCR Purification KitQiagenCatalog #28106
Uniquely Indexed Transposomes
Sci- Barcoded PCR Primers
Pitstop 2Merck MilliporeSigma (Sigma-Aldrich)Catalog #SML1169
Tween-20: working stock is 10% (100X). Aliquots are stored at 4C.
IGEPAL-630: Prepare 10% (v/v) stock made with diH20, store at Room Temperature (RT).
DAPI: Resuspend to 5 mg/mL in diH20. Aliquot and store at -20C.
Pitstop2: Resuspend in 3mM in DMSO. Aliquot and store at -20C.
Supplies List:
- 96-well PCR plates (Eppendorf, 951020427)
- 35 um cell strainer (VWR, 21008-948)
- High Sensitivity DNA Chip (Agilent, 5067-4627)
Instrument List:
- Table top centrifuge cooled to 4C with rotors for spinning 1) 96-well plates, and 2) 15 mL falcon tubes at 600 rcf
- Fluorescence Activated Cell Sorter (FACS), we use Sony SH800S
- Thermomixer with 96 well plate adapter (55C incubations at 300 rpm), we use Eppendorf Themomixer C
- Real-Time PCR instrument (Bio-Rad CFX Connect)
- DNA fluorometer or spectrophotometer (Qubit Fluorometer 2.0 is used in this protocol)
- Agilent Bioanalyzer
- Sequencing: NextSeq 500 using custom chemistry protocol
Troubleshooting
Before start
Cryopreserved tissue sections: Prepare prior to sp-sciATACseq protocol start. Refer to "Cryopreserved tissue sectioning" protocol
Uniquely indexed transposomes (8 uM): Prepare and load prior to sp-sciATACseq protocol start. Refer to "sci Transposase Loading" protocol.
Sp-sci barcoded PCR primers: Prepare prior to sp-sciATACseq protocol start. Refer to "sci Barcoded PCR Primer Preparation" protocol.
Prepare Nuclei Isolation Buffer
Construct 50mL Nuclei Isolation Buffer (NIB):
| Final Concentration | Stock Concentration | Volume of Stock | |
| 10 mM Tris HCl, pH 7.5 | 1M Tris-HCl, pH7.5 | 500 uL | |
| 10 mM NaCl | 5M NaCl | 100 uL | |
| 3mM MgCl2 | 1M MgCl2 | 150 uL | |
| 0.1 % Igepal | 10% Igepal | 500 uL | |
| 0.1 % Tween | 10% Tween | 500 uL | |
| ddH20 | to 50mL (add 48.25mL) |
Note
OPTIONAL: To prevent protease degradation, we also add 2 tablets of Pierce Preotease Inhibitor Tablets, EDTA-Free to NIB following construction. We then vortex to fully dissolve tablets.
Note
NIB is stable at 4 °C for at least 1 month without noticable degradation in library quality or nuclei dissociation ability.
Store NIB on ice throughout nuclei dissociation and preparation of tagmentation plates.
Isolate nuclei
Nuclei from cryopreserved histological sections
If sample is sourced from microbiopsy of a cryopreserved histological section, dissociate cells using NIB incubation and trituration (described below).
Note
Note
Isolation of nuclei is dependent on the sample being used. And optimization should be performed. Below we list two example nuclei isolation protocols to act as general use for cell culture and primary tissue samples. Tissue should follow a dounce homogenization protocol, while liquid cell cutures can be pelleted and resuspended directly in NIB.
This protocol is optimized for brain tissue microbiopsies. Additional optimization may need to be performed for other tissues.
1. Prepare 96-well plate(s) for microbiopsy punches
- Pipette 100 uL NIB into each well. Number of wells corresponds to number of punches to be collected.
- Seal plate and store on ice until ready to collect microbiopsies
2. Prepare instruments & tissue for collecting microbiopsies.
- Transfer cryopreserved tissue sections from -80C freezer on to dry ice in an insulated container
- Load Palkovitz punch handle with selected diameter punch (options: 250 um, 500 um, 750 um, 1 mm, 1.25 mm)
- Prechill Palkovitz punch by placing the punch in dry ice
3. Collect microbiopsies in a cryostat at -20C
- Place tissue cryosection slide in cryostat and allow ~1 min to acclimate
- Locate region of interest and collect punch
- Deposit punch in well of 96-well plate by depressing punch plunger. (Ensure that punch enters well)
- Repeat for each region to be resected. Place each new punch in new well
- Reseal 96-well plate(s)
Note
Note: Keep a record of 1) slide number, 2) punch location, and 3) well ID for each punch.
Annotating image at cryostat works well.
4. Dissociate and wash microbiopsies
- Shake plate on ice for 1 hour at 80 rpm
- Using a multi-channel pipettor, triturate each well 30x.
Note
Note: Pipette gently in order to reduce bubbles and to prevent nuclei shearing
- Spin down plate for 10 min at 500 rcf at 4C
- Using a multi-channel pipettor, aspirate 90 uL of supernatant.
Note
Note: Pellet will not be visible. Be careful to not touch sides of bottom while drawing off supernatant.
5. Dilute microbiopsy nuclei to desired concentration
Note
Note: We find that for microbiopsy punches from 200 um thick tissue /250 um biopsy punch results in (thousand nuclei):
Min: 6, 1st Q: 12, Median: 15, Mean: 16.85, 3rd Q: 22.25, Max: 29
We want 10 uL nuclei well. Each punch dissociation can be split into 4 wells (4.2K nuclei/rxn).
Therefore, we want 40 uL of 4,200 nuclei/10 uL:
C1V1 = C2V2
(1,685 nuclei/uL)(10uL) = (421.25 nuclei/uL)(x uL)
x = 40 uL
Volume to add: 40 uL - 10 uL (residual volume) = 30 uL
Final concetration of Pitstop 2 should be 70 uM in 40 uL of resuspended nuclei. Therefore:
C1V1 = C2V2
(3000 uM)(x uL) = (70 uM)(30 uL); x = 1.43 uL
Therefore, for each well, add: (1.4 uL 3 mM Pitstop 2 + 28.6 uL NIB) = 30 uL of 70 uM Pitstop 2 NIB
This should be done by making a master-mix. Given 1 plate (96 wells), prepare a master-mix for 120 wells:
(168 uL 3mM Pitstop 2 + 3,432 uL NIB) = 3600 uL of 70 uM Pitstop 2 NIB, for one plate.
- Prepare 70 uM Pitstop 2 + NIB master mix: For one plate, combine 168 uL 3 mM Pitstop2 & 3,432 uL NIB
- Add 30 uL of 70 uM Pitstop 2 NIB master mix to each well and triturate to resuspend cells
6. Split punches into multiple plates
- Split 40 uL of resuspend cells into 4 new 96-well (DNA/protein Lo-bind) plates with 10uL diluted cells/well.
Note
Note: Make sure to keep well ID consistent between plates.
96-plex Tagmentation
Prepare tagmentation plate
Add the following reagents to diluted nuclei in 96-well plate(s) (DNA and Protein Lo-bind):
Prepare 70uM Pitstop 2 + 2X TD buffer mastermix for one plate: 28 uL 3mM Pitstop2 + 1,200 uL 2X TD
Add 10 µL 70uM Pitstop 2/TD buffer (2X) to each well
Add 1 µL 8uM uniquely indexed transposase to each well
Spin down plate for 00:01:00 min at 500 rcf at 4 °C
Tagmentation
Seal plate and incubate at 55 °C with gentle shaking (300 rpm on themomixer) for 00:15:00
Place plate on ice immediately to stop reaction.
- Keep samples on ice to prevent over-transposistion and nuclei lysis.
Pool all wells for second sort
Pool all wells into 15mL conical tube, while maintaining everything on ice.
Add 2uL/per mL pooled sample of DAPI (5mg/mL) and bring to sorter for second sort.
96-plex PCR
Preparing Second Plate of Transposase Neutralization Buffer (8.5uL/well):
| Final Concentration | Stock Concentration | Volume of Stock | |
| 0.59 mg/mL | 20 mg/mL BSA | 0.25 uL | |
| 0.059% (w/v) | 1% SDS (w/v) | 0.5 uL | |
| to 8.5 uL | ddH2O | 7.75 uL |
Per well reagent volumes.
Add 2.5 uL of 10 uM i5 Indexed PCR Primer and 2.5 uL of 10 uM i7 Indexed PCR Primer prior to sort.
2nd Sort Protocol
Flow sort single nuclei based on DAPI gate
Note
Sort X nuclei per well (X is dependent on number of wells tagmented in first sort, as a linear trend)
• 96 wells (1 plate) = 22 nuclei/well for PCR
• 144 wells (1 and 1/2 plates) = 33 nuclei/well for PCR
• 192 wells (2 plates) = 44 nuclei/well for PCR etc...
Using the same gates as first sort, sort X nuclei per well into prepared second plate with modified sort settings:
- "Single cell" rather than "Normal"
This leads to a higher abort count (less efficient sorting) but is more precise in quantification
- Keep sorted samples on ice to prevent transposases cross-reacting with other nuclei.
Example gating strategy using the Sony SH800 Flow sorter:
Spin down plate at 500 rcf for 00:03:00 min at 4 °C to ensure nuclei are properly suspended in solution.
Note
Volume added, even by sorting 100 nuclei is minimal in our hands and does not require concentration adjustments.
Transposase Denaturation
Transposase Denaturation
Denature remaining transposase in sorted nuclei using SDS mixture on Eppendorf Thermocyclers.
55 °C for 00:20:00 min
Amplifying single cell libraries
Note
Nextera PCR Mater Mix currently produces the highest quality libraries. An alternative master mix using Kapa Hifi Non-Hotstart has been developed and produces good results.
Using Nextera PCR Master Mix7 steps
Add 13.5 uL PCR Master Mix to each well
7.5 µL NPM
0.25 µL 100X SYBR Green I
5.5 µL dH2O
Perform Real-time PCR on the Bio-Rad CFX Connect:
PCR protocol for Kapa Hifi Non-Hotstart Library Amplification
Pull once majority of well begin to plateau. Sci-ATAC libraries amplify between 14-22 cycles dependent on nuclei per well.
Store libraries at 4 °C for 6 months or -20 °C forever
Library Clean-up and Quantification
Pool post-PCR Product
Pool 10 uL from each well into 15mL conical tube.
Concentrate DNA via column clean up
Run full pool volume through Qiaquick PCR purification column following manufacturer's protocol.
Elute in 50 µL 10 mM Tris-HCl pH 8.0
Clean by size selection with SPRI beads
Perform a 1X SPRI bead size selection (selecting for DNA > 200 bp).
Add 50 µL 18% PEG SPRI Beads to column elution, once beads are at room temperature.
Let mixture incubate at room temperature for 00:05:00 min
Place tube on magnetic rack and wait for magnetic beads to pellet and elution to fully clear (roughly 00:02:00 min )
Remove full volume of elution without disrupting bead pellet.
Resuspend bead pellet in freshly prepared 100 µL 80% ethanol (v/v)
Remove full volume of elution without disrupting bead pellet.
Let beads fully air dry (roughly 00:08:00 min )
- Beads will first lose sheen, and then begin to form cracks.
Resuspend beads off magnetic rack in 31 µL 10 mM Tris-HCl pH 8.0
Let mixture incubate at room temperature for 00:05:00 min for DNA to fully become suspended.
Place tube on magnetic rack and wait for magnetic beads to pellet and elution to fully clear (roughly 00:20:00 min )
Remove full volume of elution without disrupting bead pellet and move to clean tube.
Qubit DNA HS Quantification
Quantify DNA concentration with 1uL eluted sample on Qubit DNA High-sensitivity kit following manufacturer's protocol.
Agilent DNA HS Bioanalyzer Quantification
Dilute sample to 4 ng/uL based on read out of Qubit by addition of 10mM Tris-HCl pH 8.0.
Run 1 uL sample on Agilent DNA HS Bioanalyzer following manufacturer's protocol.
Sequencing
Custom Nextseq500 Chemistry Protocol
Custom primers and sequencing protocol for sci-atac libraries.
