Jul 25, 2025
Version 1
Crystal structure of Enterovirus A71 2A protease mutant C110A containing VP1-2A junction in the active site apo structure (C1 PDB: 9FGO) used for fragment screen and compound soaking V.1

- Xiaomin Ni1,2,3,
- Peter Marples4,5,3,
- Daren Fearon4,5,3,
- Lizbé Koekemoer1,2,3
- 1Centre of Medicines Discovery;
- 2University of Oxford;
- 3ASAP Discovery Consortium;
- 4Diamond Light Source;
- 5Research Complex at Harwell
- Xiaomin Ni: The principle crystallographer on the 2A protease precursor project.;
- ASAP Discovery

External link: http://
Protocol Citation: Xiaomin Ni, Peter Marples, Daren Fearon, Lizbé Koekemoer 2025. Crystal structure of Enterovirus A71 2A protease mutant C110A containing VP1-2A junction in the active site apo structure (C1 PDB: 9FGO) used for fragment screen and compound soaking. protocols.io https://dx.doi.org/10.17504/protocols.io.6qpvr9b52vmk/v1
Manuscript citation:
License: This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Protocol status: Working
We use this protocol and it's working
Created: March 04, 2025
Last Modified: July 25, 2025
Protocol Integer ID: 123758
Keywords: crystallisation, XChem, ASAP, AViDD, CMD, Diamond Light Source, i04-1, Research complex at Harwell, crystal structure of enterovirus a71, crystallization of enterovirus, plasmid enterovirus coxsackievirus a71 2a protease, enterovirus a71, plasmid enterovirus coxsackievirus a71, enterovirus, 2a protease mutant c110a, hexagonal prism crystals in space group p61, crystal structure, mutant c110a, crystallization, protein, crystallization screen, hexagonal prism crystal, 2a protease, containing vp1, crystal
Funders Acknowledgements:
National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID)
Grant ID: Grant ID: U19AI171399
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgements:
Diamond Light Source Ltd, Harwell Science and Innovation Campus, Didcot OX11 0QX, UK
Research Complex at Harwell, Harwell Science and Innovation Campus, Didcot OX11 0FA, UK
Oxford Lab Technologies crystal shifter https://doi.org/10.1107/S2059798320014114
Abstract
This protocol describes the crystallization of Enterovirus 71 (EV-71) 2A protease mutant C110A containing the VP1-2A junction in the active site. The crystals form within 12-24 hours using a crystallization screen composed of 1.8M NaCl and 15% ethanol. The crystal structure was determined using X-ray diffraction, resulting in hexagonal prism crystals in space group P61 with unit cell dimensions of 61.5Å, 61.5Å, 78.9Å (90.00°, 90.00°, 120.00°) and an average resolution of 1.5Å. The protein was expressed using the plasmid Enterovirus Coxsackievirus A71 2A protease 228633.
Materials
SwissCI 3 lens crystallization plates https://swissci.com/product/3-lens-crystallisation-plate/ Codes:
Midi: UVXPO-3LENS 3W96T-PS 3W96T-UVP
1 Molarity (M) Ammonium sulfate, Molecular Dimensions, Catalog # MD2-250-35
1 Molarity (M) Sodium acetate 4.8 , Molecular Dimensions, Catalog # 133225
50% w/v PEG 2000, Molecular Dimensions, Catalog # MD2-250-17
Purified protein xxx. in 10 millimolar (mM) HEPES, 7.5 , 0.5 Molarity (M) NaCl, 5% glycerol, 0.5 millimolar (mM) TCEP
Protein construct https://www.addgene.org/204791/
Protocol materials
Enterovirus Coxsackievirus A71 inactive 2A protease with mutation C110A to preserve VP1-2A junction.addgeneCatalog #228633
Troubleshooting
Safety warnings
Follow all handling warning for the chemicals used in the crystalllisation screen composition.
Protein expression and purification
The protocol used for protein expression and purification is the following. Using the plasmid Enterovirus Coxsackievirus A71 inactive 2A protease with mutation C110A to preserve VP1-2A junction.addgeneCatalog #228633
Equipment needed
Protein sequence
ITTLGKFGQQSGAIYVGNFRVVNRHLATHNDWANLVWEDSSRDLLVSSTTAQGCDTIARCNCQTGVYYCNSMRKHYPVSFSKPSLIFVEASEYYPARYQSHLMLAVGHSEPGDAGGILRCQHGVVGIVSTGGNGLVGFADVRDLLWLDDEAMEQQ
Crystallization experiment
1d
Crystallisation screen composition:
1.8M NaCl
15% ethanol
Stock solutions used:
5 Molarity (M) Sodium chloride
100 % volume Ethanol
Note
The crystallisation screen can be stored in a duran bottle or aliquoted into 96 deep well block for easy dispensing into SwissCI 3 lens plates.
For long term storage keep the Crystallisation screen in the fridge at 4°C.
Protein and buffer requirements:
43.2 µL 15 mg/mL Sample
2.88 mL Crystallization screen
Dispense 20 µL Crystallisation screen into SwissCI 3 lens plate reservoir wells using a 100 µl multi-channel pipette.
Dispense 100 nL nl 15 mg/mL Sample to each lens using the SPT mosquito.
Dispense 100 nL Crystallisation screen to each lens using the SPT mosquito.
Drop ratio: 1:1
Final drop volume: 200 nl
Incubate at 20 °C for 24:00:00 h in Formulatrix Rock Imager.
Imaging Schedule: The first images are taken after 12 h and the imaging schedule follows a Fibonacci sequence of days for further collections.
1d
Expected result
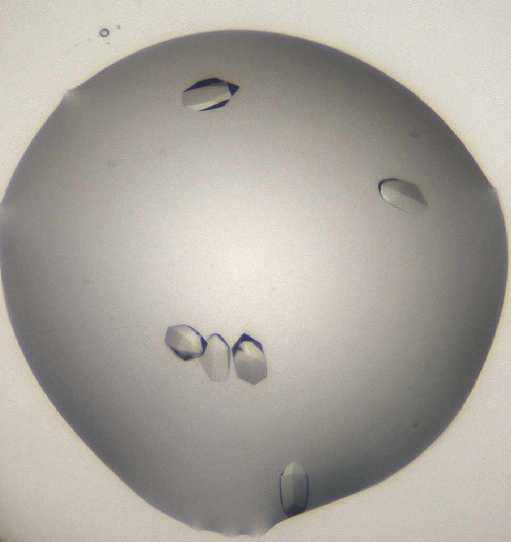
Crystals typically form within 12 h, within 24 h they have reached their maximum size. Crystals form on their own and have hexagonal prism appearance. (see image below)
Morphology:
Size: ~50 μm in width and ~100 μM in length
Average resolution: 1.5 Å
Space group: P61
Unit cell: 61.5 Å, 61.5 Å, 78.9 Å
90.00°, 90.00°, 120.00°
Data collection at Synchrotron
Diamond Light Source
Unattended Data Collection (UDC)
Data Collection Temperature: 100K
Detector: DECTRIS EIGER2 X 9M
Beamline: I04-1
Wavelength: 0.9212 Å
Resolution (Å): 1.62
Beam Size (μm): 60 X 50
Number of images: 3600
Oscillation: 0.10°
Exposure (s): 0.0020
Transmission (%): 100
Flux (ph/s): 9.50e+11
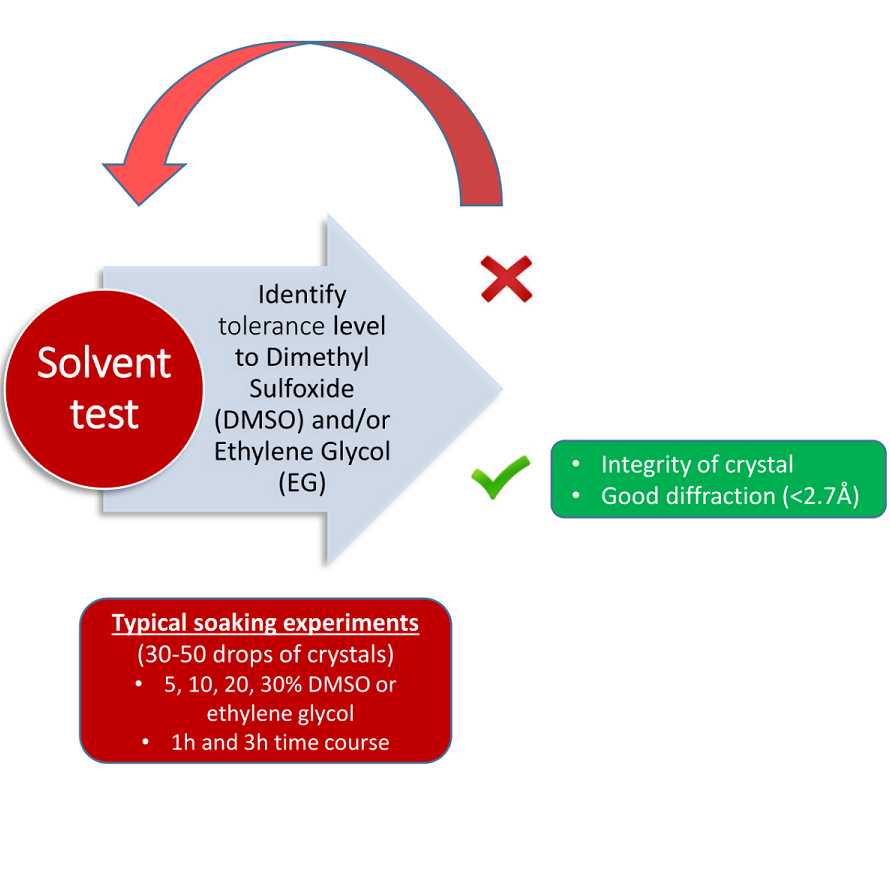
Solvent Tolerance
Fragment screen
Conditions used 5-10% DMSO, for 1h
Protocol references
N/A